-
- Downloads
Comparison Plots
Showing
- MODULES/Plotting/__pycache__/comp_plotset.cpython-38.pyc 0 additions, 0 deletionsMODULES/Plotting/__pycache__/comp_plotset.cpython-38.pyc
- MODULES/Plotting/__pycache__/general_plotset.cpython-38.pyc 0 additions, 0 deletionsMODULES/Plotting/__pycache__/general_plotset.cpython-38.pyc
- MODULES/Plotting/__pycache__/plot_settings.cpython-38.pyc 0 additions, 0 deletionsMODULES/Plotting/__pycache__/plot_settings.cpython-38.pyc
- MODULES/Plotting/comp_plotset.py 64 additions, 0 deletionsMODULES/Plotting/comp_plotset.py
- MODULES/Plotting/general_plotset.py 22 additions, 0 deletionsMODULES/Plotting/general_plotset.py
- MODULES/Plotting/obs_plotset.py 1 addition, 22 deletionsMODULES/Plotting/obs_plotset.py
- MODULES/Plotting/plot_final.py 0 additions, 0 deletionsMODULES/Plotting/plot_final.py
- MODULES/Statistics/__pycache__/data_read.cpython-38.pyc 0 additions, 0 deletionsMODULES/Statistics/__pycache__/data_read.cpython-38.pyc
- MODULES/Statistics/data_read.py 87 additions, 0 deletionsMODULES/Statistics/data_read.py
- PLOTS/ComparisonPlots/T_comparison.jpg 0 additions, 0 deletionsPLOTS/ComparisonPlots/T_comparison.jpg
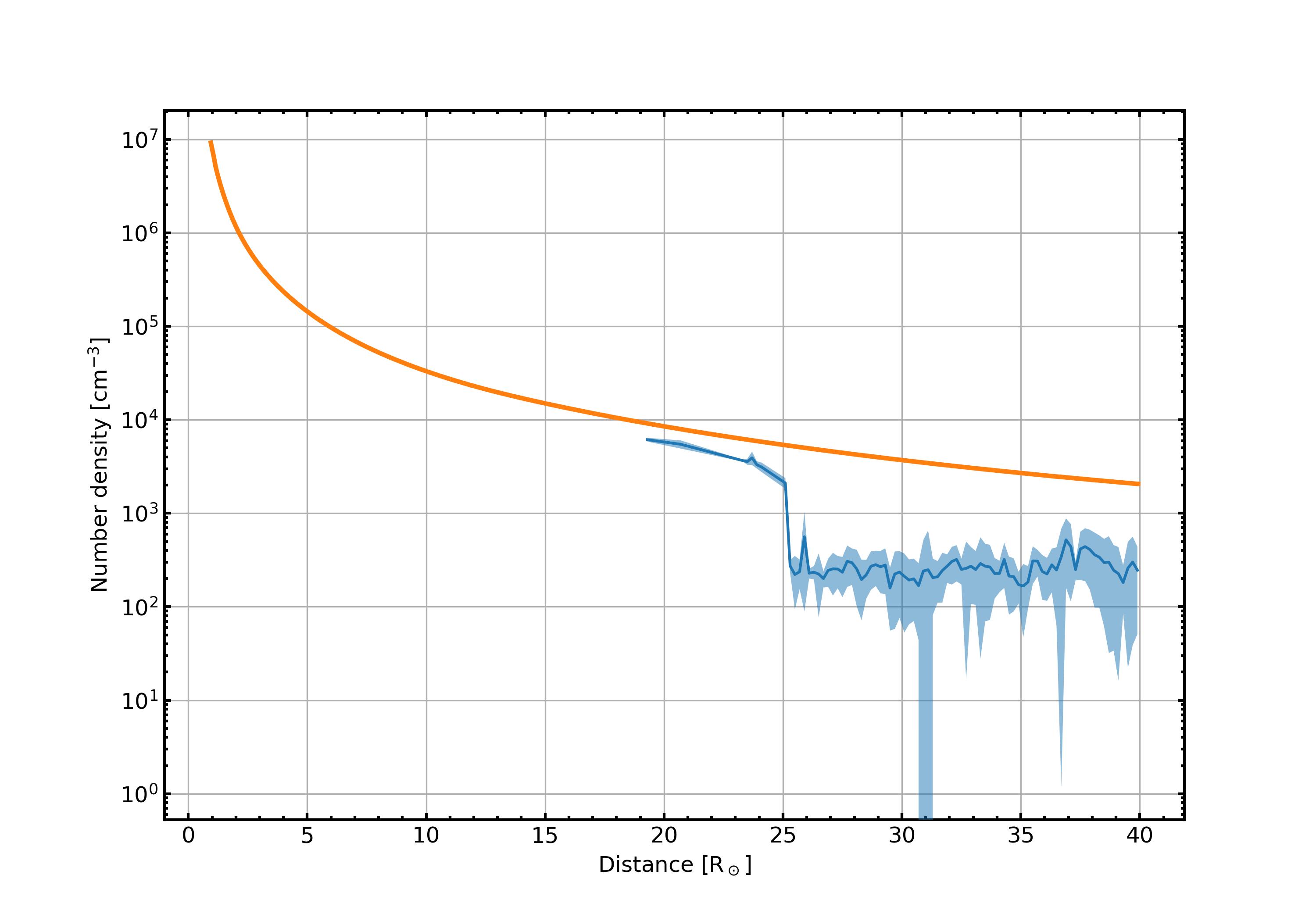
- PLOTS/ComparisonPlots/np_comparison.jpg 0 additions, 0 deletionsPLOTS/ComparisonPlots/np_comparison.jpg
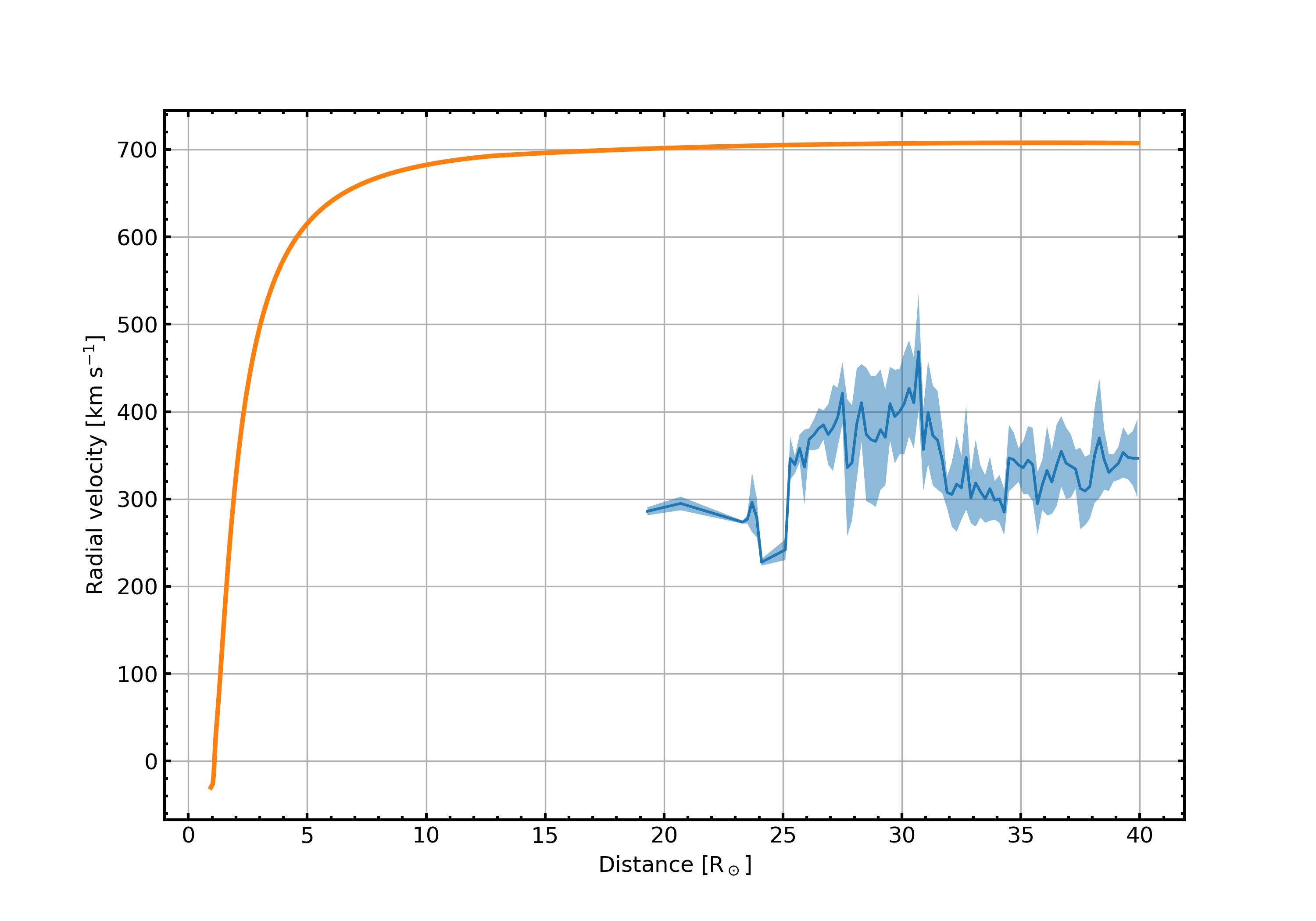
- PLOTS/ComparisonPlots/vr_comparison.jpg 0 additions, 0 deletionsPLOTS/ComparisonPlots/vr_comparison.jpg
- PLOTS/ObsDataPlots/NumberDensity_MEAN.png 0 additions, 0 deletionsPLOTS/ObsDataPlots/NumberDensity_MEAN.png
- PLOTS/ObsDataPlots/NumberDensity_MEDIAN.png 0 additions, 0 deletionsPLOTS/ObsDataPlots/NumberDensity_MEDIAN.png
- PLOTS/ObsDataPlots/RadialVelocity_MEAN.png 0 additions, 0 deletionsPLOTS/ObsDataPlots/RadialVelocity_MEAN.png
- PLOTS/ObsDataPlots/RadialVelocity_MEDIAN.png 0 additions, 0 deletionsPLOTS/ObsDataPlots/RadialVelocity_MEDIAN.png
- PLOTS/ObsDataPlots/Temperature_MEAN.png 0 additions, 0 deletionsPLOTS/ObsDataPlots/Temperature_MEAN.png
- PLOTS/ObsDataPlots/Temperature_MEDIAN.png 0 additions, 0 deletionsPLOTS/ObsDataPlots/Temperature_MEDIAN.png
- STATISTICS/sim_data.csv 1002 additions, 0 deletionsSTATISTICS/sim_data.csv
- TESTING/testing.py 1 addition, 1 deletionTESTING/testing.py
File added
File added
File deleted
MODULES/Plotting/comp_plotset.py
0 → 100644
MODULES/Plotting/general_plotset.py
0 → 100644
MODULES/Plotting/plot_final.py
0 → 100644
File added
MODULES/Statistics/data_read.py
0 → 100644
PLOTS/ComparisonPlots/T_comparison.jpg
0 → 100644
227 KiB
PLOTS/ComparisonPlots/np_comparison.jpg
0 → 100644
254 KiB
PLOTS/ComparisonPlots/vr_comparison.jpg
0 → 100644
258 KiB
File moved
File moved
File moved
File moved
File moved
STATISTICS/sim_data.csv
0 → 100644
This diff is collapsed.