-
- Downloads
wrf update
Showing
- definition-files/MPI/Singularity.miniconda3-py39-4.9.2-ubuntu-18.04-OMPI 0 additions, 89 deletions...s/MPI/Singularity.miniconda3-py39-4.9.2-ubuntu-18.04-OMPI
- definition-files/MPI/Singularity.py39.ompi 52 additions, 0 deletionsdefinition-files/MPI/Singularity.py39.ompi
- definition-files/debian/Singularity.texlive 1 addition, 1 deletiondefinition-files/debian/Singularity.texlive
- models/ICON/Singularity.gcc6 1 addition, 0 deletionsmodels/ICON/Singularity.gcc6
- models/WRF/README.md 50 additions, 0 deletionsmodels/WRF/README.md
- models/WRF/Singularity.dev 72 additions, 0 deletionsmodels/WRF/Singularity.dev
- models/WRF/Singularity.gsi 82 additions, 0 deletionsmodels/WRF/Singularity.gsi
- models/WRF/Singularity.sandbox.dev 1 addition, 1 deletionmodels/WRF/Singularity.sandbox.dev
- models/WRF/Singularity.wrf 6 additions, 0 deletionsmodels/WRF/Singularity.wrf
- models/WRF/Singularity.wrf.py 44 additions, 0 deletionsmodels/WRF/Singularity.wrf.py
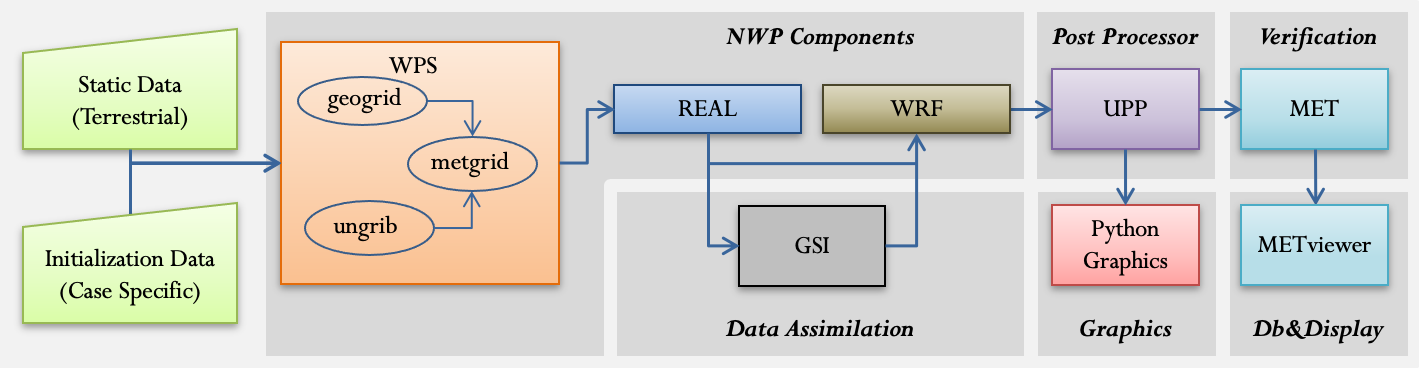
- models/WRF/dtc_nwp_flow_3.png 0 additions, 0 deletionsmodels/WRF/dtc_nwp_flow_3.png
- models/WRF/hurricane-sandy-example.sh 277 additions, 0 deletionsmodels/WRF/hurricane-sandy-example.sh
- models/WRF/hurricane-sany-example.sh 0 additions, 99 deletionsmodels/WRF/hurricane-sany-example.sh
- models/WRF/scripts/run-help 8 additions, 5 deletionsmodels/WRF/scripts/run-help
- models/WRF/scripts/run_gsi.ksh 656 additions, 0 deletionsmodels/WRF/scripts/run_gsi.ksh
- models/WRF/scripts/runscript 9 additions, 8 deletionsmodels/WRF/scripts/runscript
definition-files/MPI/Singularity.py39.ompi
0 → 100644
models/WRF/README.md
0 → 100644
models/WRF/Singularity.dev
0 → 100644
models/WRF/Singularity.gsi
0 → 100644
models/WRF/Singularity.wrf.py
0 → 100644
models/WRF/dtc_nwp_flow_3.png
0 → 100644
93.2 KiB
models/WRF/hurricane-sandy-example.sh
0 → 100755
models/WRF/hurricane-sany-example.sh
deleted
100755 → 0
models/WRF/scripts/run_gsi.ksh
0 → 100755